Demo: The unscented particle filter
Van Der Merwe, R., Doucet, A., De Freitas, N., & Wan, E. (2000). The Unscented Particle Filter. Advances In Neural Information Processing Systems, 13. Retrieved from http://papers.nips.cc/paper/1818-the-unscented-particle-filter.pdf
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.stats/Users/jhamrick/miniconda3/lib/python3.5/site-packages/matplotlib/__init__.py:872: UserWarning: axes.color_cycle is deprecated and replaced with axes.prop_cycle; please use the latter.
warnings.warn(self.msg_depr % (key, alt_key))
True model
def transition_func(x, t, omega=4*np.e-2, phi=0.5):
return 1 + np.sin(omega * np.pi * t) + phi * x
def transition(x, t, omega=4*np.e-2, phi=0.5):
vt = scipy.stats.gamma.rvs(3, scale=1/2)
return transition_func(x, t, omega=omega, phi=phi) + vt
def transition_density(xnew, xold, t, omega=4*np.e-2, phi=0.5):
xest = transition_func(xold, t, omega=omega, phi=phi)
return scipy.stats.gamma.logpdf(xnew - xest, 3, scale=1/2)
def observation_func(x, t, phi1=0.2, phi2=0.5):
if t <= 30:
return phi1 * x ** 2
else:
return phi2 * x - 2
def observation(x, t, phi1=0.2, phi2=0.5):
nt = scipy.stats.norm.rvs(loc=0, scale=np.sqrt(1e-5))
return observation_func(x, t, phi1=phi1, phi2=phi2) + nt
def observation_density(y, x, t, phi1=0.2, phi2=0.5):
yest = observation_func(x, t, phi1=phi1, phi2=phi2)
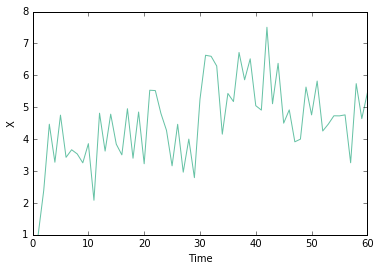
return scipy.stats.norm.logpdf(y - yest, loc=0, scale=np.sqrt(1e-5))Xorig = np.empty(60)
Xorig[0] = 1
Yorig = np.empty(60)
Yorig[0] = observation(Xorig[0], 0)
for t in range(1, 60):
Xorig[t] = transition(Xorig[t-1], t + 1)
Yorig[t] = observation(Xorig[t], t + 1)
plt.plot(np.arange(60) + 1, Xorig, '-')
plt.xlabel('Time')
plt.ylabel('X')<matplotlib.text.Text at 0x107c71278>
Regular particle filter
def particle_filter(Y, N=200):
# initialization
X = np.empty((len(Y) + 1, N))
w = np.ones((len(Y) + 1, N)) / N
for i in range(N):
X[0, i] = np.random.randn() + 1
for t in range(len(Y)):
# propagate particles and calculate unnormalized weights
Xtilde = np.empty(N)
wtilde = np.empty(N)
for i in range(N):
Xtilde[i] = transition(X[t, i], t + 1)
wtilde[i] = observation_density(Y[t], Xtilde[i], t + 1)
# normalize weights
w[t + 1] = np.exp(wtilde - scipy.misc.logsumexp(wtilde))
w[t + 1] /= np.sum(w[t + 1])
# resample particles
X[t + 1] = Xtilde[np.random.choice(np.arange(N), size=N, p=w[t + 1])]
# MCMC step
for i in range(N):
u = np.random.rand()
x = transition(X[t, i], t)
p_x = np.exp(observation_density(Y[t], x, t + 1))
p_xtilde = np.exp(observation_density(Y[t], X[t + 1, i], t + 1))
if u <= min(1, p_x / p_xtilde):
X[t + 1, i] = x
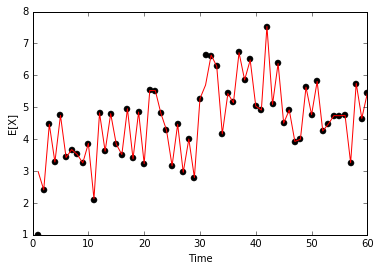
return X, wparticles = particle_filter(Yorig)
Xest = particles[0][1:].mean(axis=1)/Users/jhamrick/miniconda3/lib/python3.5/site-packages/ipykernel/__main__.py:29: RuntimeWarning: invalid value encountered in double_scalars
/Users/jhamrick/miniconda3/lib/python3.5/site-packages/ipykernel/__main__.py:29: RuntimeWarning: divide by zero encountered in double_scalars
plt.plot(np.arange(60) + 1, Xorig, 'ko')
plt.plot(np.arange(60) + 1, Xest, 'r-')
plt.xlabel('Time')
plt.ylabel('E[X]')<matplotlib.text.Text at 0x107d2ce48>
Unscented particle filter
def calc_sigma_weights(n=1, alpha=1, kappa=2, beta=0):
lam = alpha ** 2 * (n + kappa) - n
Wm = np.empty(2 * n + 1)
Wc = np.empty(2 * n + 1)
Wm[0] = lam / (n + lam)
Wc[0] = (lam / (n + lam)) + (1 - alpha ** 2 + beta)
for i in range(1, 2 * n + 1):
Wm[i] = 1 / (2 * (n + lam))
Wc[i] = 1 / (2 * (n + lam))
return Wm, Wcdef calc_sigma_points(X, P, n=1, alpha=1, kappa=2):
lam = alpha ** 2 * (n + kappa) - n
sigma = np.zeros(2 * n + 1)
sigma[0] = X.copy()
for i in range(1, n + 1):
sigma[i] = X + np.sqrt((n + lam) * P)
sigma[i + n] = X - np.sqrt((n + lam) * P)
return sigma def unscented_update(Xm_old, P_old, Y_obs, t, n=1):
# calculate sigma points and weights
Wm, Wc = calc_sigma_weights()
sigma = calc_sigma_points(Xm_old, P_old, n=n)
# propagate sigma points
X = np.zeros(2 * n + 1)
Y = np.zeros(2 * n + 1)
for i in range(2 * n + 1):
X[i] = transition(sigma[i], t)
Y[i] = observation(X[i], t)
# compute conditional t|t-1 distributions
Xm_cond = np.sum(Wm * X)
P_cond = np.sum(Wc * (X - Xm_cond) ** 2)
Ym_cond = np.sum(Wm * Y)
# incorporate new observation
Pyy = np.sum(Wc * (Y - Ym_cond) ** 2)
Pxy = np.sum(Wc * (Y - Ym_cond) * (X - Xm_cond))
K = Pxy / Pyy
Xm_new = Xm_cond + K * (Y_obs - Ym_cond)
P_new = P_cond - (K * Pyy * K)
assert P_new >= 0
return Xm_new, P_newdef propagate(Xm, P, N):
Xtilde = np.zeros(N)
for i in range(N):
Xtilde[i] = np.random.normal(Xm[i], np.sqrt(P[i]))
return Xtildedef calc_weights(Xtilde, X, Y, Xm, P, t, N):
# calculate unnormalized weights
wtilde = np.zeros(N)
for i in range(N):
p_Xtilde = scipy.stats.norm.logpdf(Xtilde[i], loc=Xm[i], scale=np.sqrt(P[i]))
p_obs = observation_density(Y, Xtilde[i], t)
p_trans = transition_density(Xtilde[i], X[i], t)
wtilde[i] = p_obs + p_trans - p_Xtilde
# normalize weights
ix = ~(np.isnan(wtilde) | np.isinf(wtilde))
w = np.zeros(N)
w[ix] = np.exp(wtilde[ix] - scipy.misc.logsumexp(wtilde[ix]))
w /= np.sum(w)
return wdef unscented_particle_filter(Y, N=200, alpha=1, beta=0, kappa=2):
# PF initialization
Y = np.concatenate([[0], Y])
X = np.zeros((len(Y), N))
w = np.ones((len(Y), N)) / N
for i in range(N):
X[0, i] = np.random.randn() + 1
# UKF initialization
Xm = np.zeros((len(Y), N))
Xm[0, :] = X[0].copy()
P = np.zeros((len(Y), N))
P[0, :] = 1
for t in range(1, len(Y)):
for i in range(N):
# perform kalman update
Xm[t, i], P[t, i] = unscented_update(Xm[t - 1, i], P[t - 1, i], Y[t], t)
# propagate particles and calculate weights
Xtilde = propagate(Xm[t], P[t], N)
wtilde = calc_weights(Xtilde, X[t - 1], Y[t - 1], Xm[t], P[t], t, N)
# resample particles
idx = np.random.choice(np.arange(N), size=N, p=wtilde)
X[t] = Xtilde[idx]
w[t] = wtilde[idx]
Xm[t] = Xm[t, idx]
P[t] = P[t, idx]
# MCMC step
for i in range(N):
u = np.random.rand()
x = transition(X[t - 1, i], t)
p_x = np.exp(observation_density(Y[t], x, t))
p_xtilde = np.exp(observation_density(Y[t], X[t, i], t))
if u <= min(1, p_x / p_xtilde):
X[t, i] = x
Xm[t, i] = x
P[t, i] = 1
return X, w, Xm, Punscented_particles = unscented_particle_filter(Yorig)
unscented_Xest = unscented_particles[0][1:].mean(axis=1)/Users/jhamrick/miniconda3/lib/python3.5/site-packages/ipykernel/__main__.py:37: RuntimeWarning: invalid value encountered in double_scalars
/Users/jhamrick/miniconda3/lib/python3.5/site-packages/ipykernel/__main__.py:37: RuntimeWarning: divide by zero encountered in double_scalars
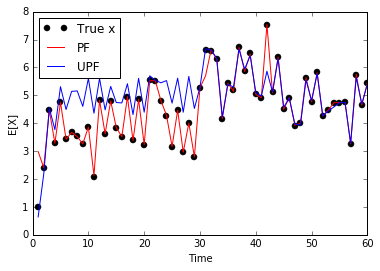
fig, ax = plt.subplots()
ax.plot(np.arange(60) + 1, Xorig, 'ko', label='True x')
ax.plot(np.arange(60) + 1, Xest, 'r-', label='PF')
ax.plot(np.arange(60) + 1, unscented_Xest, 'b-', label='UPF')
ax.set_xlabel('Time')
ax.set_ylabel('E[X]')
ax.legend(loc='best')<matplotlib.legend.Legend at 0x107fd5ac8>